Nutrient Evaluation¶
Compare nutrient fields to observations from the World Ocean Atlas. Fields included here are:
NO3
PO4
SiO3
Imports¶
%matplotlib inline
import os
import numpy as np
import xarray as xr
import utils
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import matplotlib.colors as colors
import cartopy
import cartopy.crs as ccrs
import cmocean
%load_ext watermark
%watermark -a "Mike Levy" -d -iv -m -g -h
Author: Mike Levy
Compiler : GCC 9.3.0
OS : Linux
Release : 3.10.0-1127.18.2.el7.x86_64
Machine : x86_64
Processor : x86_64
CPU cores : 72
Architecture: 64bit
Hostname: crhtc50
Git hash: 135f85d57e8712d47b21027021f8409ad12c469c
matplotlib: 3.4.2
cmocean : 2.0
cartopy : 0.19.0.post1
xarray : 0.18.2
numpy : 1.20.3
Read in the Data¶
nutrients = ['NO3', 'PO4', 'SiO3']
Read in the surface data
ds_surf_plot = xr.open_zarr('data/nutrients-surface-maps.zarr')
ds_surf_plot
<xarray.Dataset>
Dimensions: (nlat: 384, nlon: 321)
Dimensions without coordinates: nlat, nlon
Data variables:
NO3 (nlat, nlon) float32 dask.array<chunksize=(192, 321), meta=np.ndarray>
NO3_obs (nlat, nlon) float32 dask.array<chunksize=(192, 321), meta=np.ndarray>
PO4 (nlat, nlon) float32 dask.array<chunksize=(192, 321), meta=np.ndarray>
PO4_obs (nlat, nlon) float32 dask.array<chunksize=(192, 321), meta=np.ndarray>
SiO3 (nlat, nlon) float32 dask.array<chunksize=(192, 321), meta=np.ndarray>
SiO3_obs (nlat, nlon) float32 dask.array<chunksize=(192, 321), meta=np.ndarray>
TLAT (nlat, nlon) float64 dask.array<chunksize=(192, 161), meta=np.ndarray>
TLONG (nlat, nlon) float64 dask.array<chunksize=(192, 161), meta=np.ndarray>xarray.Dataset
- nlat: 384
- nlon: 321
- NO3(nlat, nlon)float32dask.array<chunksize=(192, 321), meta=np.ndarray>
- long_name :
- NO3
- units :
- mmol m$^{-3}$
Array Chunk Bytes 481.50 kiB 240.75 kiB Shape (384, 321) (192, 321) Count 3 Tasks 2 Chunks Type float32 numpy.ndarray - NO3_obs(nlat, nlon)float32dask.array<chunksize=(192, 321), meta=np.ndarray>
- long_name :
- NO3
- units :
- mmol m$^{-3}$
Array Chunk Bytes 481.50 kiB 240.75 kiB Shape (384, 321) (192, 321) Count 3 Tasks 2 Chunks Type float32 numpy.ndarray - PO4(nlat, nlon)float32dask.array<chunksize=(192, 321), meta=np.ndarray>
- long_name :
- PO4
- units :
- mmol m$^{-3}$
Array Chunk Bytes 481.50 kiB 240.75 kiB Shape (384, 321) (192, 321) Count 3 Tasks 2 Chunks Type float32 numpy.ndarray - PO4_obs(nlat, nlon)float32dask.array<chunksize=(192, 321), meta=np.ndarray>
- long_name :
- PO4
- units :
- mmol m$^{-3}$
Array Chunk Bytes 481.50 kiB 240.75 kiB Shape (384, 321) (192, 321) Count 3 Tasks 2 Chunks Type float32 numpy.ndarray - SiO3(nlat, nlon)float32dask.array<chunksize=(192, 321), meta=np.ndarray>
- long_name :
- SiO3
- units :
- mmol m$^{-3}$
Array Chunk Bytes 481.50 kiB 240.75 kiB Shape (384, 321) (192, 321) Count 3 Tasks 2 Chunks Type float32 numpy.ndarray - SiO3_obs(nlat, nlon)float32dask.array<chunksize=(192, 321), meta=np.ndarray>
- long_name :
- SiO3
- units :
- mmol m$^{-3}$
Array Chunk Bytes 481.50 kiB 240.75 kiB Shape (384, 321) (192, 321) Count 3 Tasks 2 Chunks Type float32 numpy.ndarray - TLAT(nlat, nlon)float64dask.array<chunksize=(192, 161), meta=np.ndarray>
Array Chunk Bytes 0.94 MiB 241.50 kiB Shape (384, 321) (192, 161) Count 5 Tasks 4 Chunks Type float64 numpy.ndarray - TLONG(nlat, nlon)float64dask.array<chunksize=(192, 161), meta=np.ndarray>
Array Chunk Bytes 0.94 MiB 241.50 kiB Shape (384, 321) (192, 161) Count 5 Tasks 4 Chunks Type float64 numpy.ndarray
Read in zonal mean data
ds_zonal_mean = xr.open_zarr('data/nutrients-zonal-section.zarr').compute()
for v in nutrients:
ds_zonal_mean[f'{v}_bias'] = ds_zonal_mean[v] - ds_zonal_mean[f'{v}_obs']
ds_zonal_mean = ds_zonal_mean.drop('basins').rename({'region': 'basins'})
ds_zonal_mean
<xarray.Dataset>
Dimensions: (basins: 4, lat_t: 394, z_t: 60)
Coordinates:
* lat_t (lat_t) float32 -79.22 -78.69 -78.15 -77.62 ... 88.84 89.37 89.9
* basins (basins) <U14 'Global' 'Atlantic Ocean' ... 'Indian Ocean'
* z_t (z_t) float64 5.0 15.0 25.0 ... 4.875e+03 5.125e+03 5.375e+03
z_t_ (z_t) float64 500.0 1.5e+03 2.5e+03 ... 5.125e+05 5.375e+05
Data variables:
NO3 (basins, z_t, lat_t) float32 nan nan 19.02 19.07 ... nan nan nan
NO3_obs (basins, z_t, lat_t) float32 nan nan 20.99 21.69 ... nan nan nan
PO4 (basins, z_t, lat_t) float32 nan nan 1.423 1.428 ... nan nan nan
PO4_obs (basins, z_t, lat_t) float32 nan nan 1.713 1.705 ... nan nan nan
SiO3 (basins, z_t, lat_t) float32 nan nan 42.66 43.43 ... nan nan nan
SiO3_obs (basins, z_t, lat_t) float32 nan nan 61.78 62.55 ... nan nan nan
NO3_bias (basins, z_t, lat_t) float32 nan nan -1.967 ... nan nan nan
PO4_bias (basins, z_t, lat_t) float32 nan nan -0.2902 ... nan nan nan
SiO3_bias (basins, z_t, lat_t) float32 nan nan -19.12 ... nan nan nanxarray.Dataset
- basins: 4
- lat_t: 394
- z_t: 60
- lat_t(lat_t)float32-79.22 -78.69 -78.15 ... 89.37 89.9
- edges :
- lat_t_edges
- long_name :
- Latitude Cell Centers
- units :
- degrees_north
array([-79.22052 , -78.68631 , -78.15209 , ..., 88.836334, 89.370575, 89.904816], dtype=float32) - basins(basins)<U14'Global' ... 'Indian Ocean'
array(['Global', 'Atlantic Ocean', 'Pacific Ocean', 'Indian Ocean'], dtype='<U14') - z_t(z_t)float645.0 15.0 ... 5.125e+03 5.375e+03
- long_name :
- depth from surface to midpoint of layer
- positive :
- down
- units :
- m
array([5.000000e+00, 1.500000e+01, 2.500000e+01, 3.500000e+01, 4.500000e+01, 5.500000e+01, 6.500000e+01, 7.500000e+01, 8.500000e+01, 9.500000e+01, 1.050000e+02, 1.150000e+02, 1.250000e+02, 1.350000e+02, 1.450000e+02, 1.550000e+02, 1.650984e+02, 1.754791e+02, 1.862913e+02, 1.976603e+02, 2.097114e+02, 2.225783e+02, 2.364088e+02, 2.513702e+02, 2.676542e+02, 2.854837e+02, 3.051192e+02, 3.268680e+02, 3.510935e+02, 3.782276e+02, 4.087847e+02, 4.433777e+02, 4.827367e+02, 5.277280e+02, 5.793729e+02, 6.388626e+02, 7.075633e+02, 7.870025e+02, 8.788252e+02, 9.847059e+02, 1.106204e+03, 1.244567e+03, 1.400497e+03, 1.573946e+03, 1.764003e+03, 1.968944e+03, 2.186457e+03, 2.413972e+03, 2.649001e+03, 2.889385e+03, 3.133405e+03, 3.379794e+03, 3.627670e+03, 3.876452e+03, 4.125768e+03, 4.375393e+03, 4.625190e+03, 4.875083e+03, 5.125028e+03, 5.375000e+03]) - z_t_(z_t)float64500.0 1.5e+03 ... 5.375e+05
array([5.00000000e+02, 1.50000000e+03, 2.50000000e+03, 3.50000000e+03, 4.50000000e+03, 5.50000000e+03, 6.50000000e+03, 7.50000000e+03, 8.50000000e+03, 9.50000000e+03, 1.05000000e+04, 1.15000000e+04, 1.25000000e+04, 1.35000000e+04, 1.45000000e+04, 1.55000000e+04, 1.65098398e+04, 1.75479043e+04, 1.86291270e+04, 1.97660273e+04, 2.09711387e+04, 2.22578281e+04, 2.36408828e+04, 2.51370156e+04, 2.67654199e+04, 2.85483652e+04, 3.05119219e+04, 3.26867988e+04, 3.51093477e+04, 3.78227617e+04, 4.08784648e+04, 4.43377695e+04, 4.82736719e+04, 5.27728008e+04, 5.79372891e+04, 6.38862617e+04, 7.07563281e+04, 7.87002500e+04, 8.78825234e+04, 9.84705859e+04, 1.10620422e+05, 1.24456688e+05, 1.40049719e+05, 1.57394641e+05, 1.76400328e+05, 1.96894422e+05, 2.18645656e+05, 2.41397156e+05, 2.64900125e+05, 2.88938469e+05, 3.13340469e+05, 3.37979344e+05, 3.62767031e+05, 3.87645188e+05, 4.12576812e+05, 4.37539250e+05, 4.62519031e+05, 4.87508344e+05, 5.12502812e+05, 5.37500000e+05])
- NO3(basins, z_t, lat_t)float32nan nan 19.02 19.07 ... nan nan nan
- cell_methods :
- time: mean
- grid_loc :
- 3111
- long_name :
- Dissolved Inorganic Nitrate
- units :
- mmol m$^{-3}$
array([[[ nan, nan, 19.019262 , ..., 3.127393 , 3.1124809, 3.0936654], [ nan, nan, 19.192722 , ..., 3.2082498, 3.1962583, 3.17934 ], [ nan, nan, 19.747913 , ..., 3.527372 , 3.5199492, 3.5046253], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, 21.196936 , ..., 3.127393 , 3.1124809, 3.0936654], [ nan, nan, 21.252968 , ..., 3.2082498, 3.1962583, 3.17934 ], [ nan, nan, 21.355429 , ..., 3.527372 , 3.5199492, 3.5046253], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - NO3_obs(basins, z_t, lat_t)float32nan nan 20.99 21.69 ... nan nan nan
- cell_methods :
- area: mean depth: mean time: mean within years time: mean over years
- grid_mapping :
- crs
- long_name :
- NO3
- src_file :
- /glade/work/mlevy/woa2018/1x1d/woa18_all_n00_01.nc
- src_file_varname :
- n_an
- standard_name :
- moles_concentration_of_nitrate_in_sea_water
- units :
- mmol m$^{-3}$
- wgtFile :
- /glade/work/mlevy/regrid/weight_files/latlon_1x1_180W_to_POP_gx1v7_bilinear_20180530.nc
array([[[ nan, nan, 20.986591 , ..., 0.98718345, 0.9528614 , 0.956563 ], [ nan, nan, 23.002188 , ..., 1.2074015 , 1.176309 , 1.1727661 ], [ nan, nan, 24.706089 , ..., 1.3017465 , 1.2707111 , 1.2656828 ], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, 22.391838 , ..., 0.98718345, 0.9528614 , 0.956563 ], [ nan, nan, 24.235825 , ..., 1.2074015 , 1.176309 , 1.1727661 ], [ nan, nan, 25.078253 , ..., 1.3017465 , 1.2707111 , 1.2656828 ], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - PO4(basins, z_t, lat_t)float32nan nan 1.423 1.428 ... nan nan nan
- cell_methods :
- time: mean
- grid_loc :
- 3111
- long_name :
- Dissolved Inorganic Phosphate
- units :
- mmol m$^{-3}$
array([[[ nan, nan, 1.4230961, ..., 0.9954035, 0.9968648, 0.9986534], [ nan, nan, 1.4344989, ..., 1.0013988, 1.0030755, 1.005006 ], [ nan, nan, 1.4741615, ..., 1.0225705, 1.0245651, 1.0266324], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, 1.5554473, ..., 0.9954035, 0.9968648, 0.9986534], [ nan, nan, 1.5587842, ..., 1.0013988, 1.0030755, 1.005006 ], [ nan, nan, 1.5648167, ..., 1.0225705, 1.0245651, 1.0266324], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - PO4_obs(basins, z_t, lat_t)float32nan nan 1.713 1.705 ... nan nan nan
- cell_methods :
- area: mean depth: mean time: mean within years time: mean over years
- grid_mapping :
- crs
- long_name :
- PO4
- src_file :
- /glade/work/mlevy/woa2018/1x1d/woa18_all_p00_01.nc
- src_file_varname :
- p_an
- standard_name :
- moles_concentration_of_phosphate_in_sea_water
- units :
- mmol m$^{-3}$
- wgtFile :
- /glade/work/mlevy/regrid/weight_files/latlon_1x1_180W_to_POP_gx1v7_bilinear_20180530.nc
array([[[ nan, nan, 1.7132827 , ..., 0.7824321 , 0.7851637 , 0.7851058 ], [ nan, nan, 1.8068694 , ..., 0.812785 , 0.81371224, 0.813248 ], [ nan, nan, 1.8792542 , ..., 0.75705147, 0.7578078 , 0.7565225 ], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, 1.7572992 , ..., 0.7824321 , 0.7851637 , 0.7851058 ], [ nan, nan, 1.8968039 , ..., 0.812785 , 0.81371224, 0.813248 ], [ nan, nan, 1.9512222 , ..., 0.75705147, 0.7578078 , 0.7565225 ], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - SiO3(basins, z_t, lat_t)float32nan nan 42.66 43.43 ... nan nan nan
- cell_methods :
- time: mean
- grid_loc :
- 3111
- long_name :
- Dissolved Inorganic Silicate
- units :
- mmol m$^{-3}$
array([[[ nan, nan, 42.659046 , ..., 7.488578 , 7.4984303, 7.523059 ], [ nan, nan, 42.93306 , ..., 7.5811567, 7.5947866, 7.621899 ], [ nan, nan, 43.63968 , ..., 7.896808 , 7.916356 , 7.9460306], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, 46.000652 , ..., 7.488578 , 7.4984303, 7.523059 ], [ nan, nan, 46.0551 , ..., 7.5811567, 7.5947866, 7.621899 ], [ nan, nan, 46.14845 , ..., 7.896808 , 7.916356 , 7.9460306], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - SiO3_obs(basins, z_t, lat_t)float32nan nan 61.78 62.55 ... nan nan nan
- cell_methods :
- area: mean depth: mean time: mean within years time: mean over years
- grid_mapping :
- crs
- long_name :
- SiO3
- src_file :
- /glade/work/mlevy/woa2018/1x1d/woa18_all_i00_01.nc
- src_file_varname :
- i_an
- standard_name :
- moles_concentration_of_silicate_in_sea_water
- units :
- mmol m$^{-3}$
- wgtFile :
- /glade/work/mlevy/regrid/weight_files/latlon_1x1_180W_to_POP_gx1v7_bilinear_20180530.nc
array([[[ nan, nan, 61.779434, ..., 10.102219, 10.192597, 10.225629], [ nan, nan, 63.352154, ..., 9.263759, 9.325229, 9.332197], [ nan, nan, 64.556366, ..., 9.001168, 9.07389 , 9.077654], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, 58.09269 , ..., 10.102219, 10.192597, 10.225629], [ nan, nan, 58.541718, ..., 9.263759, 9.325229, 9.332197], [ nan, nan, 60.34995 , ..., 9.001168, 9.07389 , 9.077654], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - NO3_bias(basins, z_t, lat_t)float32nan nan -1.967 ... nan nan nan
array([[[ nan, nan, -1.967329 , ..., 2.1402097, 2.1596193, 2.1371024], [ nan, nan, -3.8094654, ..., 2.0008483, 2.0199494, 2.0065737], [ nan, nan, -4.9581757, ..., 2.2256255, 2.249238 , 2.2389426], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, -1.1949024, ..., 2.1402097, 2.1596193, 2.1371024], [ nan, nan, -2.9828568, ..., 2.0008483, 2.0199494, 2.0065737], [ nan, nan, -3.722824 , ..., 2.2256255, 2.249238 , 2.2389426], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - PO4_bias(basins, z_t, lat_t)float32nan nan -0.2902 ... nan nan nan
array([[[ nan, nan, -0.29018664, ..., 0.21297145, 0.2117011 , 0.21354759], [ nan, nan, -0.37237048, ..., 0.18861377, 0.18936324, 0.19175798], [ nan, nan, -0.40509272, ..., 0.26551902, 0.2667573 , 0.27010995], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, -0.20185184, ..., 0.21297145, 0.2117011 , 0.21354759], [ nan, nan, -0.3380196 , ..., 0.18861377, 0.18936324, 0.19175798], [ nan, nan, -0.38640547, ..., 0.26551902, 0.2667573 , 0.27010995], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - SiO3_bias(basins, z_t, lat_t)float32nan nan -19.12 ... nan nan nan
array([[[ nan, nan, -19.120388 , ..., -2.6136408, -2.6941671, -2.70257 ], [ nan, nan, -20.419094 , ..., -1.6826019, -1.730442 , -1.7102981], [ nan, nan, -20.916687 , ..., -1.1043601, -1.1575336, -1.1316233], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, -12.092037 , ..., -2.6136408, -2.6941671, -2.70257 ], [ nan, nan, -12.486618 , ..., -1.6826019, -1.730442 , -1.7102981], [ nan, nan, -14.2015 , ..., -1.1043601, -1.1575336, -1.1316233], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32)
The last dataset is the global mean, with all vertical levels
glb_avg = xr.open_zarr('data/nutrients-global-profile.zarr')
glb_avg
<xarray.Dataset>
Dimensions: (z_t: 60)
Coordinates:
* z_t (z_t) float64 5.0 15.0 25.0 35.0 ... 4.875e+03 5.125e+03 5.375e+03
Data variables:
NO3 (z_t) float64 dask.array<chunksize=(60,), meta=np.ndarray>
NO3_obs (z_t) float64 dask.array<chunksize=(60,), meta=np.ndarray>
PO4 (z_t) float64 dask.array<chunksize=(60,), meta=np.ndarray>
PO4_obs (z_t) float64 dask.array<chunksize=(60,), meta=np.ndarray>
SiO3 (z_t) float64 dask.array<chunksize=(60,), meta=np.ndarray>
SiO3_obs (z_t) float64 dask.array<chunksize=(60,), meta=np.ndarray>xarray.Dataset
- z_t: 60
- z_t(z_t)float645.0 15.0 ... 5.125e+03 5.375e+03
- long_name :
- Depth
- positive :
- down
- units :
- m
array([5.000000e+00, 1.500000e+01, 2.500000e+01, 3.500000e+01, 4.500000e+01, 5.500000e+01, 6.500000e+01, 7.500000e+01, 8.500000e+01, 9.500000e+01, 1.050000e+02, 1.150000e+02, 1.250000e+02, 1.350000e+02, 1.450000e+02, 1.550000e+02, 1.650984e+02, 1.754790e+02, 1.862913e+02, 1.976603e+02, 2.097114e+02, 2.225783e+02, 2.364088e+02, 2.513702e+02, 2.676542e+02, 2.854837e+02, 3.051192e+02, 3.268680e+02, 3.510935e+02, 3.782276e+02, 4.087846e+02, 4.433777e+02, 4.827367e+02, 5.277280e+02, 5.793729e+02, 6.388626e+02, 7.075633e+02, 7.870025e+02, 8.788252e+02, 9.847059e+02, 1.106204e+03, 1.244567e+03, 1.400497e+03, 1.573946e+03, 1.764003e+03, 1.968944e+03, 2.186457e+03, 2.413972e+03, 2.649001e+03, 2.889385e+03, 3.133405e+03, 3.379793e+03, 3.627670e+03, 3.876452e+03, 4.125768e+03, 4.375392e+03, 4.625190e+03, 4.875083e+03, 5.125028e+03, 5.375000e+03])
- NO3(z_t)float64dask.array<chunksize=(60,), meta=np.ndarray>
- cell_methods :
- time: mean
- grid_loc :
- 3111
- long_name :
- NO3
- units :
- mmol m$^{-3}$
Array Chunk Bytes 480 B 480 B Shape (60,) (60,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - NO3_obs(z_t)float64dask.array<chunksize=(60,), meta=np.ndarray>
- cell_methods :
- area: mean depth: mean time: mean within years time: mean over years
- grid_mapping :
- crs
- long_name :
- NO3
- src_file :
- /glade/work/mlevy/woa2018/1x1d/woa18_all_n00_01.nc
- src_file_varname :
- n_an
- standard_name :
- moles_concentration_of_nitrate_in_sea_water
- units :
- mmol m$^{-3}$
- wgtFile :
- /glade/work/mlevy/regrid/weight_files/latlon_1x1_180W_to_POP_gx1v7_bilinear_20180530.nc
Array Chunk Bytes 480 B 480 B Shape (60,) (60,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - PO4(z_t)float64dask.array<chunksize=(60,), meta=np.ndarray>
- cell_methods :
- time: mean
- grid_loc :
- 3111
- long_name :
- PO4
- units :
- mmol m$^{-3}$
Array Chunk Bytes 480 B 480 B Shape (60,) (60,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - PO4_obs(z_t)float64dask.array<chunksize=(60,), meta=np.ndarray>
- cell_methods :
- area: mean depth: mean time: mean within years time: mean over years
- grid_mapping :
- crs
- long_name :
- PO4
- src_file :
- /glade/work/mlevy/woa2018/1x1d/woa18_all_p00_01.nc
- src_file_varname :
- p_an
- standard_name :
- moles_concentration_of_phosphate_in_sea_water
- units :
- mmol m$^{-3}$
- wgtFile :
- /glade/work/mlevy/regrid/weight_files/latlon_1x1_180W_to_POP_gx1v7_bilinear_20180530.nc
Array Chunk Bytes 480 B 480 B Shape (60,) (60,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - SiO3(z_t)float64dask.array<chunksize=(60,), meta=np.ndarray>
- cell_methods :
- time: mean
- grid_loc :
- 3111
- long_name :
- SiO3
- units :
- mmol m$^{-3}$
Array Chunk Bytes 480 B 480 B Shape (60,) (60,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - SiO3_obs(z_t)float64dask.array<chunksize=(60,), meta=np.ndarray>
- cell_methods :
- area: mean depth: mean time: mean within years time: mean over years
- grid_mapping :
- crs
- long_name :
- SiO3
- src_file :
- /glade/work/mlevy/woa2018/1x1d/woa18_all_i00_01.nc
- src_file_varname :
- i_an
- standard_name :
- moles_concentration_of_silicate_in_sea_water
- units :
- mmol m$^{-3}$
- wgtFile :
- /glade/work/mlevy/regrid/weight_files/latlon_1x1_180W_to_POP_gx1v7_bilinear_20180530.nc
Array Chunk Bytes 480 B 480 B Shape (60,) (60,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray
Plot the Output¶
We set up helper functions/classes to help with plotting!
class contour_label_format(float):
def __repr__(self):
str = '%.1f' % (self.__float__(),)
if str[-1] == '0':
return '%.0f' % self.__float__()
else:
return '%.1f' % self.__float__()
def contour_label(cl):
cl.levels = [contour_label_format(val) for val in cl.levels]
fmt = '%r'
return plt.clabel(cl, fontsize=5, inline=True, fmt=fmt)
cmap_field = cmocean.cm.dense
cmap_bias = cmocean.cm.balance
levels = dict(
NO3=[0, 0.1, 0.2, 0.4, 0.6, 0.8, 1, 2, 4, 8, 12., 16, 20, 24, 28, 32,],
PO4=[0, 0.02, 0.04, 0.08, 0.12, 0.16, 0.2, 0.3, 0.4, 0.5, 0.6, 0.8, 1.2, 1.4, 1.6, 1.8, 2.0],
SiO3=[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25, 30, 35, 40, 45, 50, 60, 70, 80, 90,]
)
def mirror_levels(v, lmax):
half = np.array(levels[v])[np.where(np.array(levels[v]) <= lmax)[0]]
return np.concatenate((-half[:0:-1], half))
levels_bias = dict(
NO3=np.arange(-12, 14, 2), #mirror_levels('NO3', 12),
PO4=np.arange(-1., 1.15, 0.15), #mirror_levels('PO4', 1),
SiO3=np.arange(-50, 55, 5), #mirror_levels('SiO3', 50),
)
contour_spec = dict(
NO3=dict(
levels=levels['NO3'],
extend='max',
cmap=cmap_field,
norm=colors.BoundaryNorm(levels['NO3'], ncolors=cmap_field.N),
),
NO3_bias=dict(
levels=levels_bias['NO3'],
extend='both',
cmap=cmap_bias,
norm=colors.BoundaryNorm(levels_bias['NO3'], ncolors=cmap_bias.N),
),
PO4=dict(
levels=levels['PO4'],
extend='max',
cmap=cmap_field,
norm=colors.BoundaryNorm(levels['PO4'], ncolors=cmap_field.N),
),
PO4_bias=dict(
levels=levels_bias['PO4'], #
extend='both',
cmap=cmap_bias,
norm=colors.BoundaryNorm(levels_bias['PO4'], ncolors=cmap_bias.N),
),
SiO3=dict(
levels=levels['SiO3'],
extend='max',
cmap=cmap_field,
norm=colors.BoundaryNorm(levels['SiO3'], ncolors=256),
),
SiO3_bias=dict(
levels=levels_bias['SiO3'], #
extend='both',
cmap=cmap_bias,
norm=colors.BoundaryNorm(levels_bias['SiO3'], ncolors=cmap_bias.N),
),
)
fig = plt.figure(figsize=(12, 8))
gs = gridspec.GridSpec(nrows=3, ncols=5, width_ratios=(1, 1, 0.02, 1, 0.02))
prj = ccrs.Robinson(central_longitude=305.0)
axs = []
maps = []
for i, nut in enumerate(['NO3', 'PO4', 'SiO3']):
ax_m = plt.subplot(gs[i, 0], projection=prj)
ax_o = plt.subplot(gs[i, 1], projection=prj)
ax_b = plt.subplot(gs[i, 3], projection=prj)
cax_field = plt.subplot(gs[i, 2])
cax_bias = plt.subplot(gs[i, 4])
axs.append((ax_m, ax_o, cax_field, ax_b, cax_bias))
maps.extend([ax_m, ax_o, ax_b])
cesm = ds_surf_plot[nut]
obs = ds_surf_plot[f'{nut}_obs']
bias = cesm - obs
units = ds_surf_plot[nut].units
cf_m = ax_m.contourf(
ds_surf_plot.TLONG, ds_surf_plot.TLAT, cesm,
**contour_spec[nut],
transform=ccrs.PlateCarree(),
)
cl_m = ax_m.contour(
ds_surf_plot.TLONG, ds_surf_plot.TLAT, cesm,
levels=contour_spec[nut]['levels'], colors='k', linewidths=0.2,
transform=ccrs.PlateCarree(),
)
#contour_label(cl_m)
cf_o = ax_o.contourf(
ds_surf_plot.TLONG, ds_surf_plot.TLAT, obs,
**contour_spec[nut],
transform=ccrs.PlateCarree(),
)
cl_o = ax_o.contour(
ds_surf_plot.TLONG, ds_surf_plot.TLAT, obs,
levels=contour_spec[nut]['levels'], colors='k', linewidths=0.2,
transform=ccrs.PlateCarree(),
)
#contour_label(cl_o)
cf_b = ax_b.contourf(
ds_surf_plot.TLONG, ds_surf_plot.TLAT, bias,
**contour_spec[f'{nut}_bias'],
transform=ccrs.PlateCarree(),
)
cl_b = ax_b.contour(
ds_surf_plot.TLONG, ds_surf_plot.TLAT, bias,
levels=contour_spec[f'{nut}_bias']['levels'], colors='k', linewidths=0.2,
transform=ccrs.PlateCarree(),
)
#contour_label(cl_b)
for ax in [ax_m, ax_o, ax_b]:
land = ax.add_feature(
cartopy.feature.NaturalEarthFeature(
'physical','land','110m',
edgecolor='face',
facecolor='gray'
)
)
ax_m.set_title(f'{nut} CESM')
ax_o.set_title(f'{nut} WOA2013')
ax_b.set_title(f'{nut} bias')
cb_field = plt.colorbar(cf_o, cax=cax_field, drawedges=True)
cb_field.ax.set_title(units)
cb_field.outline.set_linewidth(0.5)
cb_field.dividers.set_linewidth(0.25)
cb_bias = plt.colorbar(cf_b, cax=cax_bias, drawedges=True)
cb_bias.ax.set_title(units)
cb_bias.outline.set_linewidth(0.5)
cb_bias.dividers.set_linewidth(0.25)
# add a gap between left two columns and right; scale colorbars
gs.update(left=0.05, right=0.95, hspace=0.05, wspace=0.05)
offset = 0.05
cax_vert_shrink = 0.8
for i in range(len(axs)):
# the row of axis objects
ax_m, ax_o, cax_field, ax_b, cax_bias = axs[i]
# shift map
p0 = ax_b.get_position()
ax_b.set_position([p0.x0 + offset, p0.y0, p0.width, p0.height])
# shift and scale colorbar
p0 = cax_bias.get_position()
shift_up = p0.height * (1. - cax_vert_shrink) / 2
cax_bias.set_position([p0.x0 + offset, p0.y0 + shift_up, p0.width, p0.height * cax_vert_shrink])
# scale colorbar
p0 = cax_field.get_position()
shift_up = p0.height * (1. - cax_vert_shrink) / 2
cax_field.set_position([p0.x0, p0.y0 + shift_up, p0.width, p0.height * cax_vert_shrink])
utils.label_plots(fig, maps, xoff=0.02, yoff=0)
utils.savefig('nutrients-surface-maps.pdf')
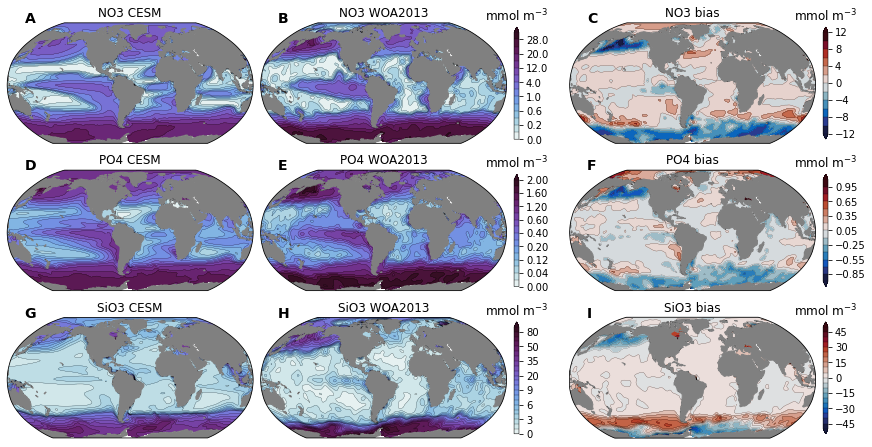
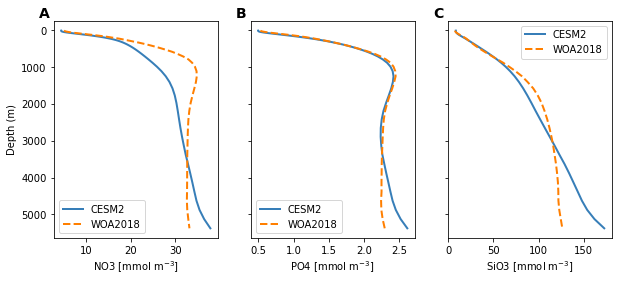
fig = plt.figure(figsize=(10, 4))
units = glb_avg.NO3.units
axs = []
for n, v in enumerate(nutrients):
ax = fig.add_subplot(1, 3, n+1)
axs.append(ax)
ax.plot(glb_avg[v], glb_avg.z_t, '-', linewidth=2, color='#377eb8', label='CESM2')
ax.plot(glb_avg[f'{v}_obs'], glb_avg.z_t, '--', linewidth=2, color='#ff7f00', label='WOA2018')
ax.set(xlabel=f'{v} [{units}]')
if n == 0:
ax.set(ylabel='Depth (m)')
else:
ax.set_yticklabels([])
plt.gca().invert_yaxis()
ax.legend()
utils.label_plots(fig, axs, xoff=-0.02, yoff=0.01)
utils.savefig('nutrients-global-profiles.pdf')
levels = dict(
NO3=np.arange(0, 51, 3),
PO4=np.arange(0, 3.8, 0.2),
SiO3=np.arange(0, 200, 10),
)
levels_bias = dict(
NO3=np.arange(-23, 26, 3),
PO4=np.arange(-1.2, 1.4, 0.2),
SiO3=np.arange(-60, 70, 10),
)
contour_spec = dict(
NO3=dict(
levels=levels['NO3'],
extend='max',
cmap=cmap_field,
norm=colors.BoundaryNorm(levels['NO3'], ncolors=cmap_field.N),
),
NO3_bias=dict(
levels=levels_bias['NO3'], #np.arange(-12, 14, 2),
extend='both',
cmap=cmap_bias,
norm=colors.BoundaryNorm(levels_bias['NO3'], ncolors=cmap_bias.N),
),
PO4=dict(
levels=levels['PO4'],
extend='max',
cmap=cmap_field,
norm=colors.BoundaryNorm(levels['PO4'], ncolors=cmap_field.N),
),
PO4_bias=dict(
levels=levels_bias['PO4'], #np.arange(-1., 1.15, 0.15),
extend='both',
cmap=cmap_bias,
norm=colors.BoundaryNorm(levels_bias['PO4'], ncolors=cmap_bias.N),
),
SiO3=dict(
levels=levels['SiO3'],
extend='max',
cmap=cmap_field,
norm=colors.BoundaryNorm(levels['SiO3'], ncolors=256),
),
SiO3_bias=dict(
levels=levels_bias['SiO3'], #np.arange(-50, 55, 5),
extend='both',
cmap=cmap_bias,
norm=colors.BoundaryNorm(levels_bias['SiO3'], ncolors=cmap_bias.N),
),
)
contour_spec.update({f'{v}_obs': contour_spec[v] for v in nutrients})
plot_basins = ['Atlantic Ocean', 'Pacific Ocean']
gss = []
col_name = ['CESM2', 'WOA2013', 'bias']
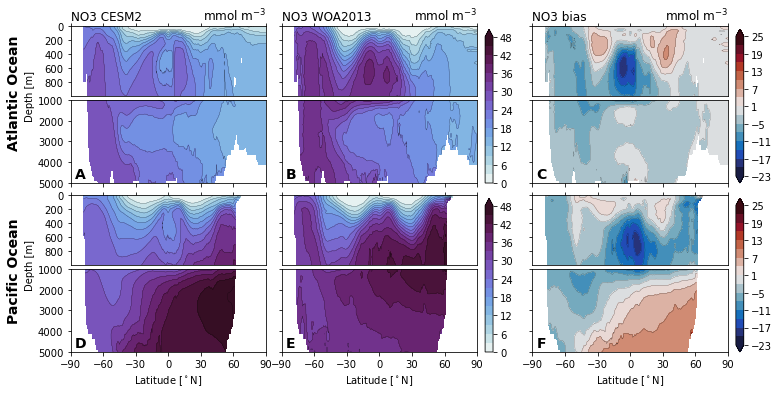
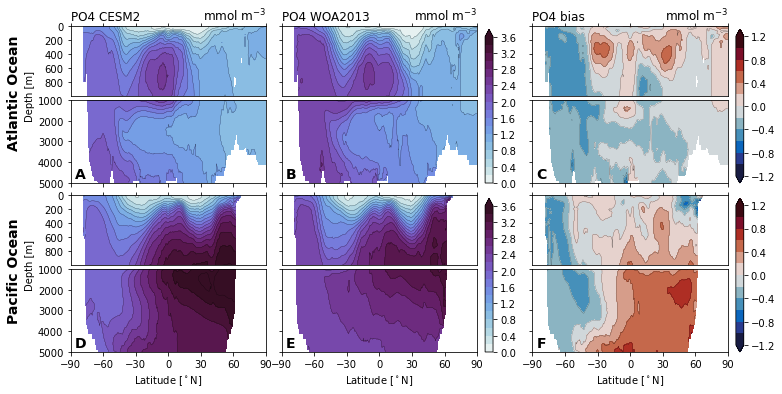
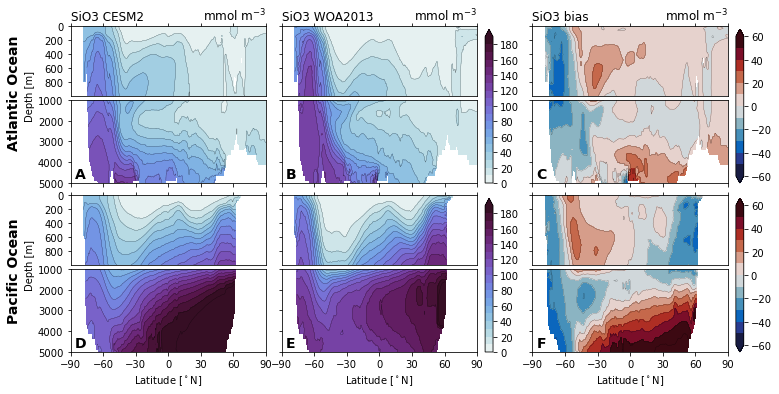
for nutrient in nutrients:
fig = plt.figure(figsize=(11, 6))
gs = gridspec.GridSpec(2, 3)
axs_surf = []
axs_deep = []
cfs_surf = []
for row, basin in enumerate(plot_basins):
axs_surf_row = []
axs_deep_row = []
cfs_surf_row = []
for col, v in enumerate([nutrient, f'{nutrient}_obs', f'{nutrient}_bias']):
gsi = gridspec.GridSpecFromSubplotSpec(100, 1, subplot_spec=gs[row, col])
ax_surf = fig.add_subplot(gsi[:45, 0])
ax_deep = fig.add_subplot(gsi[47:, 0])
axs_surf_row.append(ax_surf)
axs_deep_row.append(ax_deep)
cf = ax_surf.contourf(
ds_zonal_mean.lat_t, ds_zonal_mean.z_t,
ds_zonal_mean[v].sel(basins=basin),
**contour_spec[v]
)
cfs_surf_row.append(cf)
ax_deep.contourf(
ds_zonal_mean.lat_t, ds_zonal_mean.z_t,
ds_zonal_mean[v].sel(basins=basin),
**contour_spec[v]
)
ax_surf.contour(
ds_zonal_mean.lat_t, ds_zonal_mean.z_t,
ds_zonal_mean[v].sel(basins=basin),
levels=contour_spec[v]['levels'], colors='k', linewidths=0.2,
)
ax_deep.contour(
ds_zonal_mean.lat_t, ds_zonal_mean.z_t,
ds_zonal_mean[v].sel(basins=basin),
levels=contour_spec[v]['levels'], colors='k', linewidths=0.2,
)
ax_surf.set_ylim([1000., 0.])
ax_surf.set_yticks(np.arange(0, 1000, 200))
ax_surf.set_xticklabels([])
ax_surf.xaxis.set_ticks_position('top')
ax_surf.set_xticks(np.arange(-90, 110, 30))
ax_deep.set_ylim([5000., 1000.])
ax_deep.xaxis.set_ticks_position('bottom')
ax_deep.set_xticks(np.arange(-90, 110, 30))
if col == 0:
ax_deep.set_ylabel('Depth [m]')
ax_deep.yaxis.set_label_coords(-0.18, 1.05)
else:
ax_surf.set_yticklabels('')
ax_deep.set_yticklabels('')
if row == 1:
ax_deep.set_xlabel('Latitude [$^\circ$N]')
else:
ax_surf.set_title(f'{nutrient} {col_name[col]}', loc='left')
ax_surf.set_title('mmol m$^{-3}$', loc='right')
ax_deep.set_xticklabels('')
axs_surf.append(axs_surf_row)
axs_deep.append(axs_deep_row)
cfs_surf.append(cfs_surf_row)
gs.update(left=0.11, right=0.89, wspace=0.08,hspace=0.075)
#-- shift the right two columns over to make room for colorbar
offset = 0.05
for i in range(2):
for j in range(2, 3):
p0 = axs_surf[i][j].get_position()
axs_surf[i][j].set_position([p0.x0+offset,p0.y0,p0.width,p0.height])
p0 = axs_deep[i][j].get_position()
axs_deep[i][j].set_position([p0.x0+offset,p0.y0,p0.width,p0.height])
#-- add colorbars
for i in range(2):
for j in [1, 2]:
p0 = axs_surf[i][j].get_position()
p1 = axs_deep[i][j].get_position()
cbaxes = fig.add_axes([p1.x0 + p1.width + 0.01,
p1.y0 + 0.0,
0.01,
p0.height + p1.height - 0.0])
cb = fig.colorbar(cfs_surf[i][j], cax=cbaxes)
fig.text(0.03, 0.6, plot_basins[0],
fontsize=14.,
fontweight = 'semibold',rotation=90);
fig.text(0.03, 0.2, plot_basins[1],
fontsize=14.,
fontweight = 'semibold',rotation=90);
utils.label_plots(fig, [ax for ax_row in axs_deep for ax in ax_row] , xoff=0.005, yoff=-0.18)
utils.savefig(f'nutrients-sections-{nutrient}.pdf')