Running the model#
Setup a feisty integration.
%load_ext autoreload
%autoreload 2
import matplotlib.pyplot as plt
import feisty
Configure testcase to run#
settings_in = dict()
settings_in["benthic_prey"] = {
"defaults": {"benthic_efficiency": 0.075, "carrying_capacity": 0},
"members": [{"name": "benthic_prey"}],
}
settings_in["food_web"] = [
{"predator": "Sf", "prey": "Zoo", "preference": 1.0},
{"predator": "Sp", "prey": "Zoo", "preference": 1.0},
{"predator": "Sd", "prey": "Zoo", "preference": 1.0},
{"predator": "Mf", "prey": "Zoo", "preference": 0.45},
{"predator": "Mf", "prey": "Sf", "preference": 1.0},
{"predator": "Mf", "prey": "Sp", "preference": 1.0},
{"predator": "Mf", "prey": "Sd", "preference": 1.0},
{"predator": "Mp", "prey": "Zoo", "preference": 0.45},
{"predator": "Mp", "prey": "Sf", "preference": 1.0},
{"predator": "Mp", "prey": "Sp", "preference": 1.0},
{"predator": "Mp", "prey": "Sd", "preference": 1.0},
{"predator": "Md", "prey": "benthic_prey", "preference": 1.0},
{"predator": "Lp", "prey": "Mf", "preference": 0.5},
{"predator": "Lp", "prey": "Mp", "preference": 1.0},
{"predator": "Lp", "prey": "Md", "preference": 1.0},
{"predator": "Ld", "prey": "Mf", "preference": 0.375},
{"predator": "Ld", "prey": "Mp", "preference": 0.75},
{"predator": "Ld", "prey": "Md", "preference": 1.0},
{"predator": "Ld", "prey": "benthic_prey", "preference": 1.0},
]
Run the model#
ds_out = feisty.config_and_run_testcase(
"tanh_shelf",
"cyclic",
start_date='0001-01-01',
end_date='0002-12-31',
settings_in=settings_in,
diagnostic_names=[
"T_habitat",
"ingestion_rate",
"predation_flux",
"predation_rate",
"metabolism_rate",
"mortality_rate",
"energy_avail_rate",
"growth_rate",
"reproduction_rate",
"recruitment_flux",
"fish_catch_rate",
"encounter_rate_link",
"encounter_rate_total",
"consumption_rate_max_pred",
"consumption_rate_link",
],
)
Starting template generation at 15:32:39
Starting run() at 15:32:40
Integrating 730 steps (starting at 15:32:40)
Starting a new output dataset for timestep 365 (15:32:50)
Finished _solve at 15:32:58
Output#
ds_out.info()
xarray.Dataset {
dimensions:
X = 22 ;
group = 9 ;
time = 730 ;
fish = 8 ;
feeding_link = 19 ;
variables:
float64 X(X) ;
<U12 group(group) ;
float64 biomass(time, group, X) ;
<U2 fish(fish) ;
<U15 feeding_link(feeding_link) ;
<U2 predator(feeding_link) ;
<U12 prey(feeding_link) ;
float64 T_habitat(time, fish, X) ;
float64 ingestion_rate(time, fish, X) ;
float64 predation_flux(time, fish, X) ;
float64 predation_rate(time, fish, X) ;
float64 metabolism_rate(time, fish, X) ;
float64 mortality_rate(time, fish, X) ;
float64 energy_avail_rate(time, fish, X) ;
float64 growth_rate(time, fish, X) ;
float64 reproduction_rate(time, fish, X) ;
float64 recruitment_flux(time, fish, X) ;
float64 fish_catch_rate(time, fish, X) ;
float64 encounter_rate_link(time, feeding_link, X) ;
float64 encounter_rate_total(time, fish, X) ;
float64 consumption_rate_max_pred(time, fish, X) ;
float64 consumption_rate_link(time, feeding_link, X) ;
object time(time) ;
// global attributes:
}
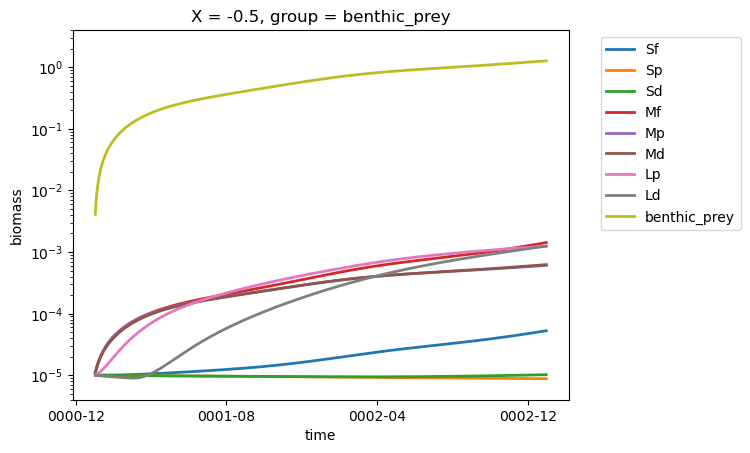
test_ds = ds_out.isel(X=0, time=0)
for group in test_ds.group.data:
ds_out.biomass.sel(group=group).isel(X=0).plot(linewidth=2)
plt.ylim([4e-6, 4])
plt.yscale("log")
plt.legend(test_ds.group.data, bbox_to_anchor=(1.05, 1.0));